# Check out the help page for this dataset
help(GBSG2, package = "TH.data")
# Load the data
data(GBSG2, package = "TH.data")
# Look at the summary of the dataset
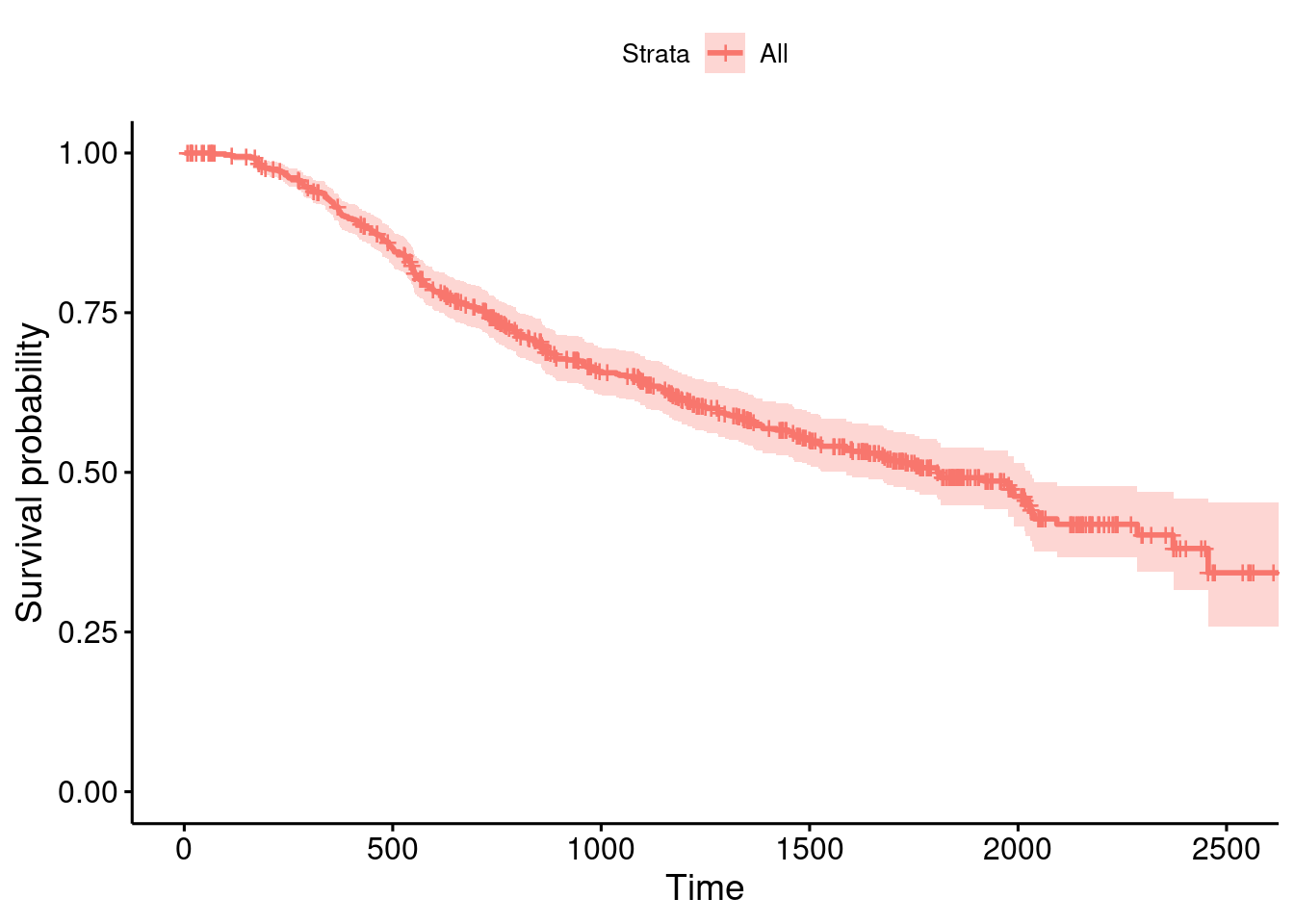
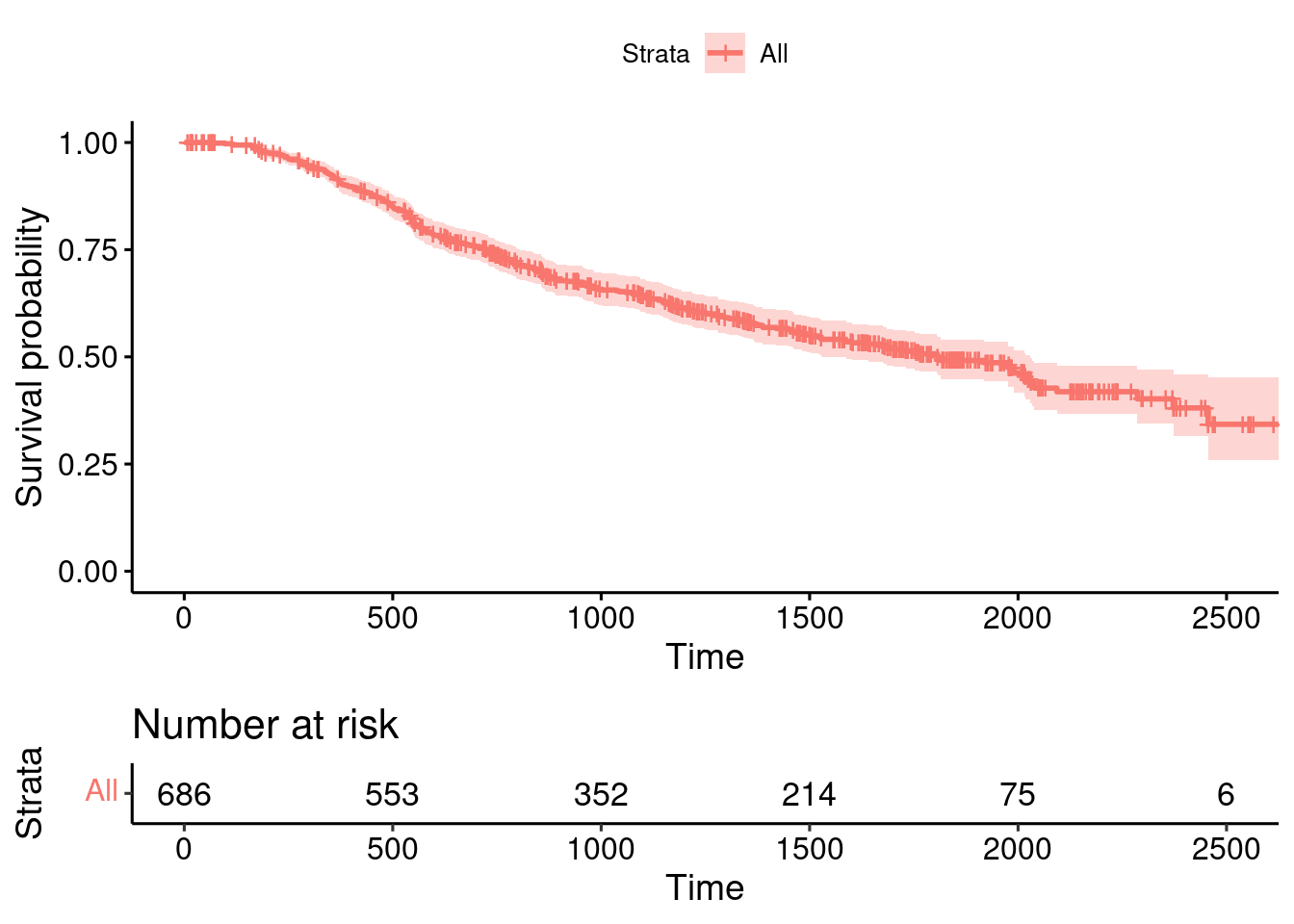
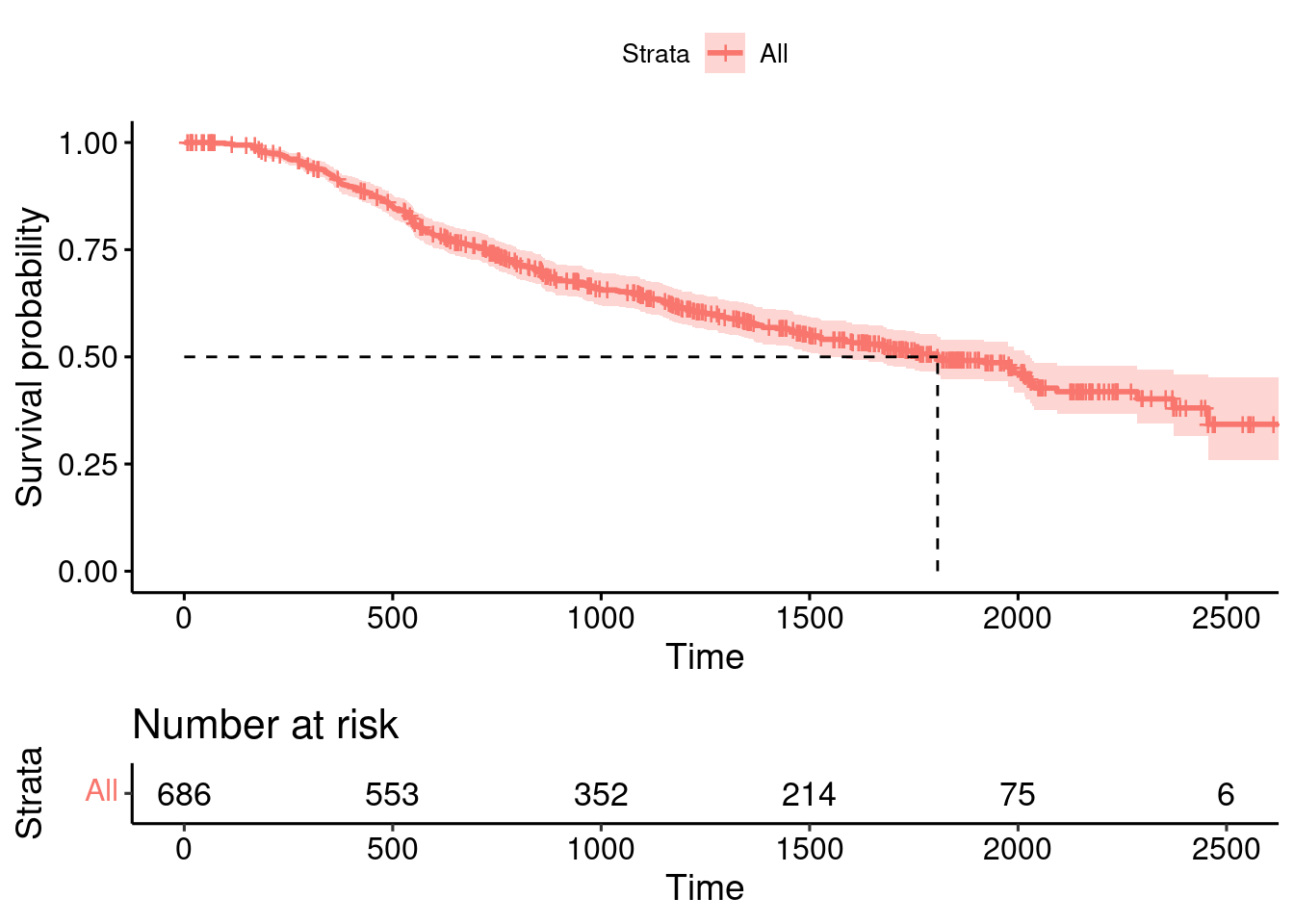
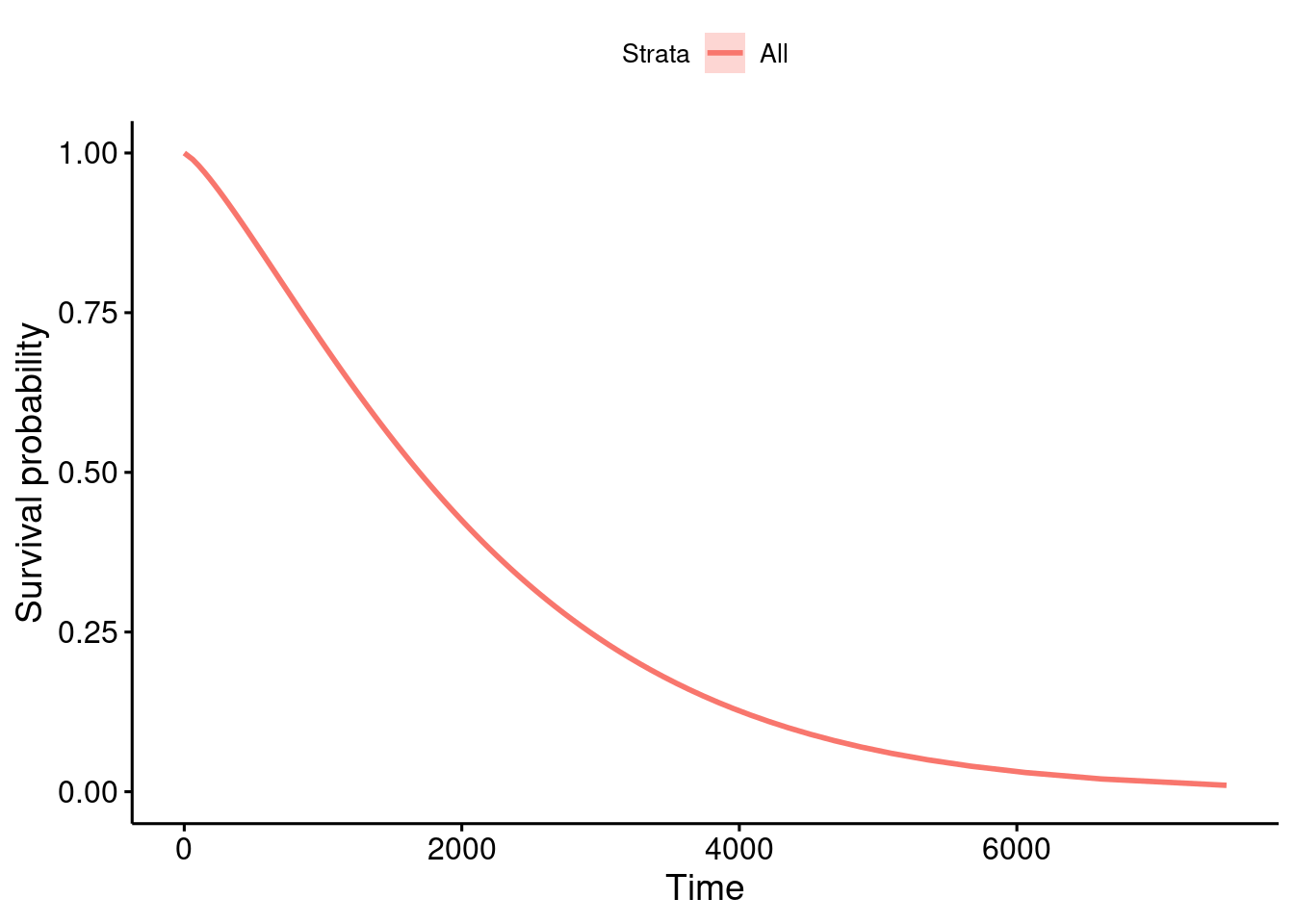
summary(GBSG2) horTh age menostat tsize tgrade
no :440 Min. :21.00 Pre :290 Min. : 3.00 I : 81
yes:246 1st Qu.:46.00 Post:396 1st Qu.: 20.00 II :444
Median :53.00 Median : 25.00 III:161
Mean :53.05 Mean : 29.33
3rd Qu.:61.00 3rd Qu.: 35.00
Max. :80.00 Max. :120.00
pnodes progrec estrec time
Min. : 1.00 Min. : 0.0 Min. : 0.00 Min. : 8.0
1st Qu.: 1.00 1st Qu.: 7.0 1st Qu.: 8.00 1st Qu.: 567.8
Median : 3.00 Median : 32.5 Median : 36.00 Median :1084.0
Mean : 5.01 Mean : 110.0 Mean : 96.25 Mean :1124.5
3rd Qu.: 7.00 3rd Qu.: 131.8 3rd Qu.: 114.00 3rd Qu.:1684.8
Max. :51.00 Max. :2380.0 Max. :1144.00 Max. :2659.0
cens
Min. :0.0000
1st Qu.:0.0000
Median :0.0000
Mean :0.4359
3rd Qu.:1.0000
Max. :1.0000